Document Type : Original Article
Authors
- Yoslaine Ruiz 1
- Natacha Carlos 1
- Alexis Musacchio 2
- Yadira Sanchez 1
- Danay Callard 1
- Rosabel Perez 1
- Merardo Pujol 1
- Alejandro Fuentes 1
1 Department of Plant Biotechnology, Center for Genetic Engineering and Biotechnology, Ave. 31/190 and 158, Playa, P.O. Box 6162. Havana 10600, Cuba.
2 Department of System Biology, Center for Genetic Engineering and Biotechnology, Ave. 31/190 and 158, Playa, P.O. Box 6162. Havana 10600, Cuba
Abstract
Bean golden yellow mosaic virus (BGYMV) is considered the main drawback for bean production in the Caribbean basin and South Florida, where it is widely distributed. In Cuba, BGYMV disease has been reported along the entire territory and several BGYMV isolates were described in western Cuba. Now, further prospection uncovers a new BGYMV isolate in Ciego de Avila province in the center of the island, which together with the previous isolates configured a broader epidemiological picture of the country. An in-depth analysis of BGYMV genome sequences revealed many single nucleotide polymorphisms contrasting the sequences of isolates found in Cuba and those outside the island, albeit their phylogenetic closeness. An analysis of viral protein sequences disclose specific amino acids in the coat, replication enhancer, nuclear shuttle (NSP) proteins, distinguishing the new isolate from that of western Cuba and from the continental consensuses. Moreover, other amino acid substitutions were predicted in the replication associated protein (Rep) and movement proteins. The protein modeling discloses some new amino acid interactions that together with previous findings suggest a gap in gene/amino-acid sequences when compared the isolates in Cuba and those of the continent. Interesting, the transcriptional activator protein (TrAP) is the most conservative protein in all analyzed BGYMV isolates being totally conservative. Thus, with the invariable Rep, the TrAP could be considered as suitable supports to build molecular antiviral strategies in a way to protect local bean cultivation.
Graphical Abstract
Keywords
Main Subjects
- Introduction
|
*Corresponding Author: Alejandro Fuentes([email protected]) |
Gnomes [1]. The genomes of bipartite begomoviruses consist of two separate circular single-stranded DNA (DNA-A and DNA-B), whereas monopartite exhibits only one single-stranded DNA [1]. Interestingly, the genus Begomovirus is the largest one, which groups viruses that affect many important crops such cassava, tomato, pepper, and bean among others.
The common bean (Phaseolus vulgaris L.), a staple crop in Cuba, has been affected by several begomoviruses [2, 3]. Importantly, Bean golden yellow mosaic virus (BGYMV) is considered as the main drawback for bean production in the island [2, 3]. Moreover, BGYMV has been recognized as endemic and it is one of the most serious viral diseases of the common bean in the Caribbean basin and South Florida [4, 5]. Stunted and distorted plant growth and a striking green-yellow mosaic in the leaves are the main symptoms described in bean plants infected with BGYMV [6, 7]. Consequently, yield losses can reach 100 % of the cultivated plants when they are infected during the early stages of growth. The same symptom patterns produced by BGYMV were described in bean plants infected with the begomovirus Bean golden mosaic virus (BGMV) albeit in Brazil and Argentina [8]. BGYMV differs from BGMV in its genome DNA sequence; BGMV is phloem-restricted in beans, while BGYMV is transmissible through the sap to the bean germplasm [9]. Further investigations in Caribbean basin verified many BGYMV isolates that exhibited similar symptoms, and also several single nucleotide inconsistences, suggesting an ongoing variation of this species.
The BGYMV genome is bipartite. The DNA-A encompasses the open reading frames (ORFs) required for replication (AC1/Rep and AC3/REn), transcriptional regulation (AC2/TrAP), and encapsidation (AV1/CP), while the DNA-B includes ORFs responsible for short and long distance movements; BV1 (NSP) is involved in the movement of viral DNA from the nucleus to the cytoplasm, whereas BC1 (MP) mediates cell-to-cell movement across the cell wall via the plasmodesmata. Both A and B components have a high similar non-coding common region (CR) containing the TAATATTAC sequence, which is the origin of the virion strand replication [10] and repeated sequences TGGAG (known as “iterons”) that are binding sequences of the replication-associated protein (Rep) [11].
The previous analysis on the BGYMV genome demonstrated the large similarities between the isolates found in western Cuba and those of other Caribbean basin areas. Now, a new isolate from the Ciego de Avila province (in the central part of Cuba) was cloned and characterized, enriching the list of local BGYMV variants. The present BGYMV genome analysis of local isolates and comparison with those described in the Caribbean basin reveals single nucleotide polymorphisms along the virus genomes. In correspondence, the amino acid substitutions and their interactions suggest a dissociation of BGYMV isolates found in Cuba from the continental/regional relatives and there is a probable evolutionary gap separating them. Thus, the new BGYMV isolate from Ciego de Avila appeared to be closer to the local BGYMV variants, which strengthen the postulation of the island population of BGYMV variants.
- Materials and Methods
2.1. Origin of symptomatic bean leaves
A leaf sample from symptomatic common bean plant (Phaseolus vulgaris), showing typical yellow mosaic symptoms, was collected in a production area of the Ciego de Avila province (in the center of Cuba) and used for the virus isolation.
2.2. Cloning the BGYMV genome and sequence analysis
Total plant DNA was extracted from a plant leaf sample by the CTAB method [12]. Detection of the geminiviral genome was conducted by PCR by using degenerated primers PAL1v1978/PAR1c496 [13]. The amplified fragments were cloned in pGEM–T-easy vector system (Promega) and sequenced. The sequence comparison was made by using BLAST EMBL tools.
For viral genome magnification by rolling circle amplification (RCA) a φ29 polymerase was used according to the kit instructions of Ilustra Templiphi 100 (GE Healthcare Life Sciences) with few modifications described previously [14]. The amplicons were digested with the HindIII and BglII enzymes for the linearization of the viral genome and resolved in 0.8 % agarose gel. The digested amplicons were cloned in a pBluescriptIISK (+) vector (Stratagene) and sequenced. Two independent recombinant clones of component A and two of B were obtained.
The identity analysis of the cloned sequences was performed by using BLASTN (www.ncbi.nlm.nih.gov). The phylogenetic analyses were based on the full-length sequences of begomoviruses found infecting bean plants in the Caribbean basin areas, available in NCBI-GenBank database, and listed by the Geminiviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV). Multiple sequence alignments were obtained by using the MUSCLE algorithm [15] implemented in MEGA7 [16] and edited manually.
The phylogenetic relationships were represented in trees constructed by using Maximum Likelihood, considering the corrected Akaike Information Criterion (AICc) and Bayesian Information Criterion (BIC). The phylogenetic tree for DNA-A components of BGYMV and for the begomoviruses identified in bean plants was generated by the GTR+G nucleotide substitution model. The tree for DNA-B sequences was constructed by using the JC+G+I model. Each case was bootstrapped with 1000 replicates.
2.3. Construction of infective BGYMV clones and replication assays in Nicotiana benthamiana and bean plants
A pBluescriptIISK(+) A vector, including a copy of the BGYMV A component, was digested with HindIII and BglII enzymes. The fragment obtained (999 bp) was cloned in pCAMBIA3300. An additional complete unit of BGYMV A (2644 bp) was generated from pBluescriptIISK(+) A digested with HindIII and cloned into pCAMBIA3300, containing the previous BGYMV fragment (999 bp). The resulting pCAMBIA3300 BGYMV A hemi-dimer construction contained a complete viral genome unit of the BGYMV component A, delimited at each end by common regions (CR) arranged in tandem.
A pBluescriptIISK(+) B vector, containing a copy of the BGYMV B component, was digested with HindIII and SmaI enzymes. The resulting fragment (2141 bp) was cloned in pCAMBIA3300. An additional complete unit of BGYMV B (2608 pb) was generated from pBluescriptIISK(+) B digested with HindIII and cloned into pCAMBIA3300, containing the previous B component fragment (2141 pb). The resulting pCAMBIA3300 BGYMV B hemi-dimer construction contained a complete viral genome unit of BGYMV component B, delimited at each end by common regions (CR) arranged in tandem.
Binary vectors pCAMBIA3300 BGYMV A and pCAMBIA3300 BGYMV B were mobilized into an Agrobacterium tumefaciens strain LBA4404 by the freeze-thaw method. The adaxial leaf surface of the fourth-leaf stage Nicotiana benthamiana plants were agro-infiltrated with a mix of equal volumes of the Agrobacterium tumefaciens clones prepared as described [14]. In beans, seven-day-old seedlings were agro-inoculated in the cut along the elongating epicotyls as described [7] with the above-mentioned bacterial preparation. Agro-inoculated plants were maintained in a controlled environment for plant cultivation under the biosafety license LH47-L (119) of Centro Nacional de Seguridad Biológica (National Biological Safety Center), Cuba.
2.4. Southern blot hybridization
Total DNA from the top leaves of Nicotiana benthamiana and bean plants was purified 15 days after inoculation, following the CTAB method [12]. For Southern blot hybridization, samples of total DNA (1 µg) were resolved in a 0.8 % agarose gel and transferred to Hybond N+ (GE Healthcare, Amersham) after denaturation by using capillary blotting. After blotting, the transferred DNA was fixed to the membrane by using an Ultraviolet Crosslinker Device (Amersham Pharmacia Biotech). The hybridization was performed at 60 oC by using the [α-32P]dATP-labeled common region (CR) as a probe. The probe was prepared by the random Primer-Gene Labeling System (Promega). The hybridization procedure was followed as described in the Hybond N+ manufacturer’s instructions. The exposure of radiolabeled membranes was carried out on Lucky films (China lucky film corporation).
2.5. Nucleotide sequence accession numbers
The genome sequences corresponding to components A and B of the BGYMV isolate from Ciego de Avila were deposited in GenBank under access numbers MW082628 and MW082629, respectively.
2.6. Predicting amino-acid sequence and structural modeling of viral proteins
The amino acid sequences of BGYMV ORFs were predicted by using bioinformatic tools available at the Vector NTI Software (Invitrogen). The 3D structure models of CP (AV1), Rep (AC1), TrAP (AC2), REn (AC3), NSP (BV1), and MP (BC1) proteins were constructed by using the SWISS–MODEL tools [17], W252–W258, https://doi.org/10.1093/nar/gku340) and the Robetta services for structural predictions are from the Berkeley Open Infrastructure for Network Computing Platform [18], (Web Server issue): W526-31. doi: 10.1093/nar/gkh468. PMID: 15215442; PMCID: PMC441606). The contact-protein interaction analysis of modeled proteins was performed by using the interactive chemical computing and molecular modeling tool MOE (Molecular Operating Environment (MOE), 2014.09, Chemical Computing Group Inc., 1010 Sherbooke St. Western, Suite #910, Montreal, QC, Canada, H3A 2R7, 2014).
- Results
3.1. Identification and sequence analysis of BGYMV on bean plants grown in Ciego de Avila province, demonstration of its relatedness to the western Cuba isolates.
A leaf of a common bean plant showing golden yellowing was sampled during a field survey of farming areas of Ciego de Avila province, in the center of Cuba. The total DNA was amplified via PCR by using the degenerated primers described above. The resulting amplicons of nearly 1100 bp were cloned and sequenced. As a result of the sequence analysis and through a comparison with the available genome sequences, it was found that three DNA amplicons showed 99 %, 99 %, and 97 % identity to BGYMV A, previously characterized [3]. It was therefore concluded that the symptomatic bean plant sample was infected with BGYMV.
The total DNA from the plant sample was exposed to rolling circle amplification (RCA) to further trace BGYMV. The RCA product was digested with HindIII and BglII restriction enzymes. As expected, the digested RCA product was resolved as a single band of dsDNA of viral genome size in agarose gel. The fragment (nearly 3 kb) was cloned in pBluescriptIISK (+) and sequenced. The analysis of two recombinant clones revealed 2644 bp inserts, corresponding to the A component (MW082628) that shares 98 % identity with the BGYMV-Cu isolates from western Cuba (KX185518, KU160634, KX185517, and AJ544531). The other two recombinant clones showed 2608 bp inserts, corresponding to the B component (MW082629), that shares 94% to 97% nucleotide identities with the GenBank reference genomes of western Cuba, Caribbean basin, and South Florida (KU145406, DQ119825, AF173556, LO1636, M91605, and M10080).
From the ICTV and GenBank databases, the BGYMV type isolates in each country and other virus species identified in common bean in the Caribbean area were used for the phylogenetic relationship creation. All BGYMV components A were grouped in the same cluster (Figure 1a). The component A of BGYMV Ciego de Avila isolate is close to other isolates from western Cuba (particularly to the KX185517), and are arranged in the same branch. In the nearest branch is that BGYMV isolate located in Florida, (DQ119824), with a very small evolutionary distance, corresponding to the geographical proximity. The same cluster associates the isolates from Guatemala (M91604), the Dominican Republic (L01635), and Puerto Rico (M10070), located in another branch, and the isolate from Mexico (AF173555). The begomoviruses Common bean mottle virus (KX011473) and Common bean severe mosaic virus (KX011476), isolated in Cuba [2], were not clustered with BGYMV isolates which support their phylogenetic dissociation.

Figure 1. Phylogenetic trees based on full-length genome sequences of BGYMV isolates and the begomoviruses identified in bean plants. a: phylogenetic tree of genome component A, built by using the Maximum Likelihood method based on the GTR+G model. b: phylogenetic tree of genome component B, built by using the Maximum Likelihood method based on the JC+G+I model. Bootstrap score percentage of 1000 replicates are indicated at each node. The scale bars indicate the nucleotide substitutions per site. The genome sequences of African cassava mosaic virus was included as an out group
The B component of the BGYMV Ciego de Avila isolate is closer to the other B component isolated in western Cuba (KU145406) (Figure 1b). It is also grouped together with those of Florida (DQ119825), Mexico (AF173556), the Dominican Republic (L01636), and Guatemala (M91605); the short length of the branch to which they belong, indicates high similarities. The B component isolated in Puerto Rico (M10080) is located in a different branch, is more distant. Other begomoviruses identified in beans such as Bean dwarf mosaic virus [Colombia] (M88180), Bean chlorotic mosaic virus [Venezuela] (JQ283246) and Bean chlorosis virus [Venezuela] (JN848771) have been clustered separately from the new B component, like their A components.
The genome sequence of the BGYMV Ciego de Avila isolate was exhaustively compared with a consensus of BGYMV genomes found in western Cuba (local) and with the consensus of isolates representing the Caribbean basin and South Florida (regional or continental). These comparisons unveiled several SNPs scattered along components A and B. The SNPs were more abundant in component B, 92 in regard to the regional consensus and 60 in regard to the local consensus, in the A component we found 90 SNPs in regard to the regional consensus, and 25 SNPs in regard to the local consensus (Figure 2 a,b).
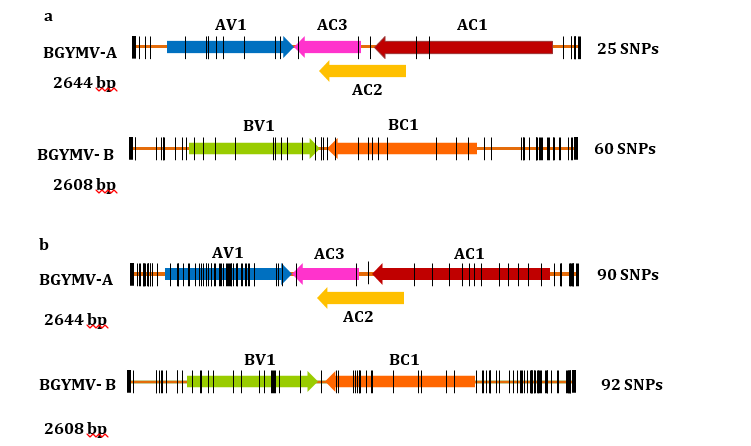
Figure 2. Representation of single nucleotide polymorphisms (SNPs) in BGYMV components of the Ciego de Avila isolate compared with western Cuba and continental consensuses. a: SNPs in regard to western Cuba consensus. b: SNPs with regard to the regional consensus
It is noteworthy that most SNPs in the new component A, compared with the local consensus, were situated in the CR (11) and in the AV1 (CP) (9) gene sequences. Another two SNPs were found alongside AC3 (REn) and AC2 (one matching in AC3) (Figure 2a). As a result of these SNPs, two amino acids were predicted to be substituted in CP (A36S and V81T) and two others in the REn protein (G6E and E126V) (Figure 3a), which denotes that most SNPs in component A were silent. Interestingly, the same two amino acid substitutions, predicted for REn and CP in the new isolate in relation to the local consensus, were found as compared with the regional consensus (Figure 3b). Moreover, a high similarity was found in the CP, TrAP, and REn amino acid sequences between local and regional consensuses in relation to the new isolate (Figure 3 a,b), which may suggest a common origin. Considering the global small number of SNPs with respect to the local consensus, we can plausibly postulate that the new component A emerged from resident variants, whereas the regional consensus of component A recedes, outnumbering the SNPs in the AV1 gene (44), followed by CR (30) and AC1 (13)
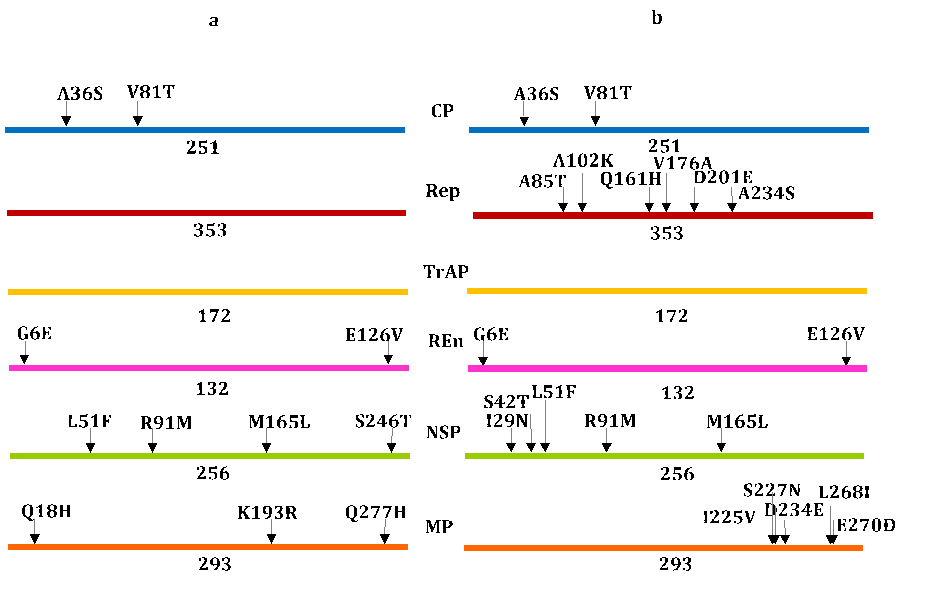
Figure 3. Representation of amino acid substitutions in BGYMV proteins of the isolate from Ciego de Avila in comparison to western Cuba consensus (a) and continental consensus (b). The amino acids in each substitution are expressed by using one code letter
In further support to the above assumption, the comparison of the new isolate with the local consensus reveals only two SNPs in the AC1 gene that were silent. Meanwhile, more SNPs were found in the AC1 gene in respect to the regional consensus, foreseeing six amino acid substitutions (A85T, A102K, Q161H, V176A, D201E, and A234S) (Figure 3b), which stresses the dissociation between the Cuban and regional/continental variants. One sustenance for these dissociations could be the competition between viruses during co-infections [19, 20] in more assorted environments of the continent.
In the case of component B, the SNPs were explored in the new Ciego de Avila isolate in regard to the only component B recorded in western Cuba (KU145406) and the consensus of component B of the regional isolates analyzed earlier. With respect to isolate KU145406, the analysis reveals several SNPs in the CR (36), BV1 gene (11), BC1 gene (10), and another three in the region between the 3´ end of BV1 and BC1 open reading frames (Figure 2a). These SNPs match four amino acids substitutions in the NSP protein (L51F, R91M, M165L, and S246T) and three in the MP protein (Q18H, K193R, and Q277H) (Figure 3a). With regard to the regional consensus, the intergenic region alone reaches 59 SNPs, while BV1 and BC1 genes accumulated 16 each, and one more was found between the 3´ ends of the BV1 -BC1 open reading frames (Figure 2b). As a result of these SNPs, five amino acid substitutions (I29N, S42T, L51F, R91M, and M165L) scattered along the NSP protein were deduced, whereas another five (I225V, S227N, D234E, L268I, and E270D) were set up in the MP protein (Figure 3b).
3.2. Viability analysis for BGYMV isolate from Ciego de Avila
Infective clones of both A and B genomic components were developed to assess the capacity of BGYMV isolated in Ciego de Avila in replication and movement. They included one functional unit of each viral DNA component. The functional units comprise two viral intergenic regions tandemly flanking the viral genome copy (Figure 4 b, d) that enables viral replication by the rolling circle replication mechanism.
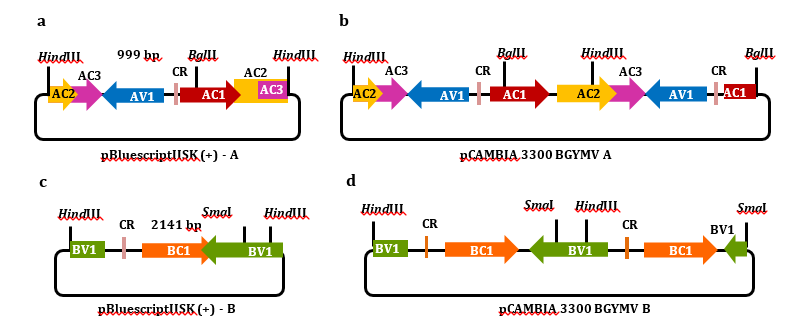
Figure 4. Constructions of cloned BGYMV A and B genome components in the pBluescriptIISK(+) (a and c) and their hemi-dimers in pCAMBIA 3300 (b and d), respectively. The open reading frames are represented as AC1 (Rep), AC2 (TrAP), AC3 (REn), AV1 (CP), BC1 (NSP), and BV1 (MP). The common region is represented as CR. The restriction enzymes HindIII, BglII, and SmaI used to generate constructions are indicated
The infection process was monitored by symptom tracking and viral replicative forms were sensed by the Southern blot hybridization assay, using CR as probe. As a result of this study, the golden mosaic symptoms appeared in common bean plants 15 days post-inoculation, correlating with the corresponding BGYMV replicative forms (Figure 5, left). In the case of Nicotiana benthamiana plants, symptoms did not appear at all albeit the abundant viral replicative forms were probed (Figure 5, right). Both results supported the notion of functionality/viability of the new isolate of BGYMV.
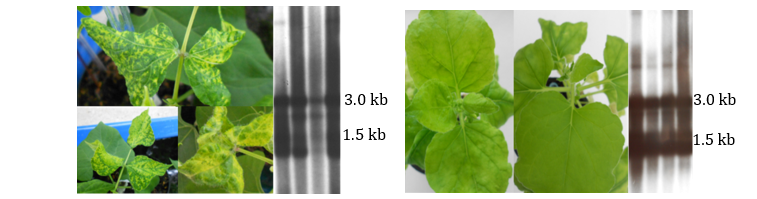
Figure 5. BGYMV viability assay. Left and right correspond to bean and Nicotiana benthamiana plants inoculated with the BGYMV isolate of Ciego de Avila, respectively. Bean leaves show typical golden yellowing. Each plant illustration is accompanied with the corresponding autoradiography of Southern blot assay detecting BGYMV DNA; the major replicative forms are indicated.
3.3. Structural modeling of BGYMV proteins corresponding to Ciego de Avila isolate compared with consensuses of local and continental isolates
Structural 3D models for each BGYMV protein sequence were constructed to further explore whether the predicted amino acid substitutions could have an impact on amino acid interactions along the protein structure. Furthermore, structural domains and functional motifs described by other authors for begomovirus proteins were considered for this analysis.
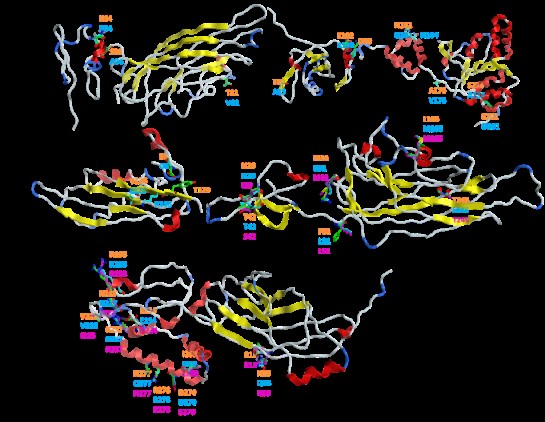
In the CP protein, the A36S substitution does not change its interaction with N34. The other substitution, i.e. V81T, exposed in a protein loop, maintains the same non-interacting status with the amino acids in its vicinity. The S36 is situated in the initial α-helices of the protein and does not affect the nuclear/nucleolar localization signal (NLSs) sequences in the N-terminal end of the protein, presented here as 16KVSRS20 and related to the previously described arginine-rich cluster, 16KVRRR20 [21]. The motif located in the central region of the protein, presented here as 46RKPR49 and related to the previously described 52RKPR55, was not affected either. The minimal transmission domains between 123-149 amino acids and the nuclear export signal (NES) located at the C-terminal end [21] of the protein was not affected by amino acid substitutions. Thus, it is considered that the CP functions are highly conserved among the analyzed isolates and no consequences of the amino acid changes should be expected.
Rep is of particular interest; it is essential for rolling circle replication (RCR) [22]. Its multifunctional nature and the several domains it combines [23] could make it especially sensitive to amino acid substitutions. No amino acid differences have been established between the local consensus and the Ciego de Avila BGYMV isolate, which expresses their high proximity and common origin. A different picture was obtained with respect to the continental consensus; where among the substituted amino acids (a total of six) K102 interacted with D99 by hydrogen bound. With the substitution of Q161H, the hydrogen bound to N164 is lost. The A85T, V176A, D201E, and A234S substitutions seemed not to be involved in new interactions. All substitutions were distributed along all four protein domains: A85T and A102K in the DNA nicking and binding domains, Q161H and V176A in oligomerization domain as well as D201E and A234S in the helicase domain [23]. None of them were interrupting the functional motifs I, II, III, Walker A, Walker B, B’, and C [24-26]. From the presented data, it is not predictable that whether these amino acids changes would have any impact on Rep activities. Nevertheless, these Rep analyses strengthen the idea of evolutionary dissociations between the BGYMV isolates of the island and the continent.
REn protein has been described to interact with itself, Rep, and PCNA (proliferating cell nuclear antigen) [27]. Hydrophobic residues in the middle of the REn protein seem to be involved in those interactions, while polar residues in both N and C-terminals of the protein are important for the interaction of REn with the retinoblastoma-related protein (pRBR) [27]. These interactions are considered to be involved in replication and infection processes. The 3D model for the REn protein reveals four α-helices (between amino acids 10-16, 52-58, 79-97, and 101-115), one more than that previously described (56-65, 79-95, and 101-116) [28] . The G6E substitution (at the N-terminal end) produced a new hydrogen bond with T129 and the E126V substitution (at the C-terminal end) lost the associated K128, previously interacting with hydrogen and ion bonds. These changes do not affect the α-helix structures described above. The amino acids E6 and V126 are specific for the BGYMV isolate of Ciego de Avila and could be considered one of the marks of the new REn variant.
Two nuclear localization sequence (NLS) motifs, structured in NLS-A (residues 27 to 39, R(L/T)S(A/I)(N/V)KRHDGKRR) and NLS-B (residues 81 to 96, (S/Q)(K/L)GKMEPNR(S/C)RSYIKL) were described mediate nuclear import of the NSP protein [29, 30]. In NLS-A, the I29 was substituted with N29, as compared with the regional consensus (I29N). In NLS-B, the R91 of the continental consensus was substituted with M91. Hence, we may hypothesize that these changes could have an impact on protein nuclear import activity. For the other amino acid substitutions, S42T, L51F and M165L, no effects can be anticipated, since new interactions were not envisaged. Interestingly, the L51F, R91M and M165L substitutions are common from local and continental protein consensus. The F51, M91, and L165 amino acids may distinguish the NSP protein of the new BGYMV isolate.
Two protein domains, the “anchor domain” (amino acids 117 to 180) and the “pilot domain” (amino acids 1 to 49) mediate the intracellular sorting of the MP protein [31]. In the 3D model, the K193R and Q277H substitutions with respect to the local consensus and those of I225V, S227N, D234E, L268I, and E270D with regard to the regional consensus do not disturb the overall interaction picture. However, the interaction of Q18 with R19 was lost. Whether the Q18H could affect the pilot domain function, still needs to be studied.
It is noteworthy that the TrAP protein appeared to be the most conservative among all BGYMV variants.
It should be mentioned that only two amino acids, and in rare cases three, were found supplanting each site. This probably, occurred because of protein structure constraints or functional requirements limiting the diversity of the involved amino acids. The new attributes, predicted as a result of the above described amino acid changes that withstand a specific structure or functionalities of proteins may include the following: (i) The replacement of methyl by hydroxyl protons and vice versa, providing or withdrawing the hydrogen binding potential. (ii) The addition of methyl groups provoking ionic environment. (iii) Aromatic stacking by the imidazole ring, which could be neutral or positively charged depending on the microenvironment. (iv) The elongating side chain with methylene groups crowned with amine nitrogen or carboxylic acid groups that are charged and interacting with hydrogen and ionic bonds. (v) Changing the negatively charged carboxylic groups with amid group, retaining the high polarity of the structure, and (vi) Increasing the positive charge by the guanidine group or being an effective donor of methylene by the sulfide group in the side chain [32]. In addition, in many cases, amino acids could serve as substrates for kinases and glycosyltransferases.
- Discussion
A new variant of BGYMV from Ciego de Avila emerged as an isolate that is phylogenetically closer to BGYMV isolates in western Cuba as well as to isolates from the Caribbean basin and South Florida. In spite of this, it exhibited many SNPs. The SNPs in the intergenic region point to the divergence between the isolates of western Cuba and the one from Ciego de Avila which is probably a consequence of self-mutational processes, originating just after the isolate arrived from the continent. The CR mutation rate is higher than that of the coding regions, suggesting that the CR is subjected to less-selective constraints, which reinforces the notion on the CR as a variable region in geminiviruses [33]. The complete identity between the AC1 sequences of the Ciego de Avila isolate with the consensus of those of western Cuba and its dissociation from the continental consensus, suggests a common origin of the Cuban BGYMV variants. Interestingly, the new isolate exhibited particular amino acid hallmarks in the CP (S36 and T81) and REn (E6 and V126) proteins that distinguish component A from other isolates. At the same time, amino acids F51, M91, and L165 in the NSP protein could be considered as a specific feature of the new BGYMV B component. Furthermore, the additional Rep, NSP, and MP protein dissociations between the continental and the local variants suggest that an evolutionary margin between both geographical areas may exist. As indicated, the proportion of amino acid changes in relation to the number of proteins in component B is higher than in component A with respect to consensuses, local, and continental. Component A has overlapping genes with multifaceted functions that should preserved, including transactivation, Rep-iteron recognition, Rep-REn interaction, and CP-vector interaction [21] that may restrain its mutational rate and may explain this behavior. In the case of component B, it could also be assumed as a separate module in certain circumstances that facilitates its exchange between different isolates and may magnify its adaptation/dissemination and its variability [21] over component A. Thus, it can hypothesize that amino acid substitutions may have at least two origins: single nucleotide mutations [21] and component B interchanges between BGYMV located within and outside the island. The predicted 3D models for viral proteins revealed few new amino acid interactions that did not appear to have a marked impact on virus infectivity and disclosed some flexibility of the BGYMV genome/proteome.
Nucleotide changes in the new BGYMV isolate are not unusual. Quasispecies were frequently described in begomovirus populations emerging in in vivo and in vitro infected plants as a consequence of ensembles of recombinant and mutated viral genomes [34, 35]. Besides, the mutation rate of begomovirus resembles that of RNA viruses of which the RNA dependent RNA polymerase exhibits a mutation frequency of 10-3 -10-4 per nucleotide, as a consequence of it poor fidelity [36]. However, since geminivirus DNA replication occurs in the nucleus, where the belief of a higher fidelity of host DNA polymerases leads to the assumption of less viral DNA population variation, it could be assumed that other cell processes are further involved in extending DNA mutations, for example, during mixed infections. It is therefore likely that during the replication processes, the host can drive the emergence and fitness of new begomovirus quasispecies [34]. It is not clear which virus-associated mechanisms are activated during the replication of a particular geminivirus in a particular host or environment affecting the fidelity of the replication processes. However, it is of particular interest how many disassociations of BGYMV genomes could be found in a small island like Cuba, or how much dissimilarity can be accumulated in BGYMV genomes in the nearby surrounding regions of the American continent where plant and vector diversity can boost virus speciation. From the above data, it is justifiable to speculate that the new BGYMV variant is not independent of the previous BGYMV collection in the island; it probably rose in bean cultivation systems or wild reservoirs where at least two populations of BGYMV alternatives could be hypothesized for the moment, here described in western Cuba and the one of Ciego de Avila. It is noteworthy that this variant of BGYMV is highly functional and it suggests that it probably emerged as a viable entity, confirming the quasispecies theory [36]. Accordingly, the analysis presented here on BGYMV genome dynamics contributed to further distinguish those evolutionary margins and drips between the island and the continent, supporting quasispecies emergence.
Studies to enhance host antiviral mechanisms, by targeting specific viral transcripts/genes of begomoviruses, were successfully described in various plant species by using molecular designs to potentiate plant RNA silencing [37]. Thus, assessing previous results, we could assume that similar strategies could be suitable to protect beans from the effects of local BGYMV, where the main targets could be the transcripts of Rep and TrAP proteins. Thereby, we accept that both are scarcely variable proteins/genes in the local scenario and that they offer several irreplaceable functions for the virus cycle, for which reason we adopt that they could be suitable supports for building conjoint antiviral strategies.
Conflicts of interest
The authors declare that they have no known conflicts of interest that could have influenced the work described in this paper.
Consent for publications
All authors and the corresponding authorities approved the final manuscript for publication.
Availability of data and material
Data are available on request from the authors.
Authors' contributions
YR performed research, analyzed data, and contributed to the draft; NC contributed to data analysis and processing, and contributed to the draft; AM analyzed the data and contributed new protein models; YS contributed methods and research; DC contributed methods and research; RP contributed research; MP conceived the study; AF conceived and designed the experiments, analyzed data, and wrote the paper. All authors read and contributed to the final version of the manuscript.
Funding
It is not applicable.
Ethics approval and consent to participate
The authors declare that the presented study was performed without involving Human Participants and/or Animals.
Acknowledgements
This work was supported by the Center for Genetic Engineering and Biotechnology, projects 3031-237 and 3031-500. The authors would like to thank Dr. Miriam Ribas for her help in English language editing.
Orcid
Yoslaine Ruiz: https://www.orcid.org/0000-0001-9453-2558
Merardo Pujol: https://www.orcid.org/0000-0002-5429-7095
Alejandro Fuentes: https://www.orcid.org/0000-0003-3468-2436
- Fiallo-Olivé E, Lett J-M, Martin D P, Roumagnac P, Varsani A, Zerbini F M, Navas-Castillo J, Consortium I R. (2021). ICTV virus taxonomy profile: Geminiviridae 2021. The Journal of General Virology, 102(12): 001696 [Crossref], [Google Scholar], [Publisher]
- Chang-Sidorchuk L, González-Alvarez H, Navas-Castillo J, Fiallo-Olivé E, Martínez-Zubiaur Y. (2017). Complete genome sequences of two novel bipartite begomoviruses infecting common bean in Cuba. Archives of virology, 162(5): 1431-1433. [Crossref], [Google Scholar], [Publisher]
- Echemendía A, Ramos P, Peral R, Fuentes A, González G, Sanpedro J, Morales F. (2001). Cuban isolate of Bean golden yellow mosaic virus is a member of the Mesoamerican BGYMV group. Plant Disease, 85(9): 1030-1030. [Crossref], [Google Scholar], [Publisher]
- Morales F J. (2006). History and current distribution of begomoviruses in Latin America. Advances in virus research, 67: 127-162. [Crossref], [Google Scholar], [Publisher]
- Blair M, Bassett M, Abouzid A, Hiebert E, Polston J, McMillan Jr R, Graves W, Lamberts M. (1995). Occurrence of bean golden mosaic virus in Florida. Plant Disease, 79(5): 529-533. [Crossref], [Google Scholar], [Publisher]
- Blair M W, Beaver J S, Nin J C, Prophete E, Singh S P. (2006). Registration of PR9745-232 and RMC-3 red-mottled dry bean germplasm lines with resistance to Bean golden yellow mosaic virus. Crop science, 46(2): 1000. [Crossref], [Google Scholar], [Publisher]
- Garrido-Ramirez E R, Sudarshana M R, Gilbertson R L. (2000). Bean golden yellow mosaic virus from Chiapas, Mexico: Characterization, Pseudorecombination with Other Bean-Infecting Geminiviruses and Germ Plasm Screening. Phytopathology, 90(11): 1224-1232. [Crossref], [Google Scholar], [Publisher]
- Gilbertson R L, Faria J C, Ahlquist P, Maxwell D P. (1993). Genetic diversity in geminiviruses causing bean golden mosaic disease: the nucleotide sequence of the infectious cloned DNA components of a Brazilian isolate of bean golden mosaic geminivirus. Phytopathology-New York and Baltimore Then St Paul-, 83: 709-709. [Crossref], [Google Scholar], [Publisher]
- Morra M R, Petty I T. (2000). Tissue specificity of geminivirus infection is genetically determined. The Plant Cell, 12(11): 2259-2270. [Crossref], [Google Scholar], [Publisher]
- Hanley-Bowdoin L, Settlage S B, Orozco B M, Nagar S, Robertson D. (1999). Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Critical Reviews in Plant Sciences, 18(1): 71-106. [Crossref], [Google Scholar], [Publisher]
- Argüello-Astorga G, Ruiz-Medrano R. (2001). An iteron-related domain is associated to Motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Archives of virology, 146(8): 1465-1485. [Crossref], [Google Scholar], [Publisher]
- Doyle J J, Doyle J L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12(13): 39-40. [Google Scholar], [Publisher]
- Rojas M, Gilbertson R, Maxwell D. (1993). Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant disease, 77(4): 340-347. [Crossref], [Google Scholar], [Publisher]
- Fuentes A, Carlos N, Ruiz Y, Callard D, Sánchez Y, Ochagavía M E, Seguin J, Malpica-López N, Hohn T, Lecca M R. (2016). Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Molecular Plant-Microbe Interactions, 29(3): 197-209. [Crossref], [Google Scholar], [Publisher]
- Edgar R C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research, 32(5): 1792-1797. [Crossref], [Google Scholar], [Publisher]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution, 33(7): 1870-1874. [Crossref], [Google Scholar], [Publisher]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino T G, Bertoni M, Bordoli L. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research, 42(W1): W252-W258. [Crossref], [Google Scholar], [Publisher]
- Kim D E, Chivian D, Baker D. (2004). Protein structure prediction and analysis using the Robetta server. Nucleic acids research, 32(suppl_2): W526-W531. [Crossref], [Google Scholar], [Publisher]
- Ali A, Roossinck M J. (2017). Analysis of quasispecies variation in single and mixed viral infection. Virus evolution, 3(2): vex037. [Crossref], [Google Scholar], [Publisher]
- Roossinck M J. (2017). Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus research, 239: 82-86. [Crossref], [Google Scholar], [Publisher]
- Sharma P, Ikegami M. (2009). Characterization of signals that dictate nuclear/nucleolar and cytoplasmic shuttling of the capsid protein of Tomato leaf curl Java virus associated with DNAβ satellite. Virus research, 144(1-2): 145-153. [Crossref], [Google Scholar], [Publisher]
- Elmer J S, Brand L, Sunter G, Gardiner W E, Bisaro D M, Rogers S G. (1988). Genetic analysis of the tomato golden mosaic virus II. The product of the AL1 coding sequence is required for replication. Nucleic acids research, 16(14): 7043-7060. [Crossref], [Google Scholar], [Publisher]
- Fondong V N. (2013). Geminivirus protein structure and function. Molecular plant pathology, 14(6): 635-649. [Crossref], [Google Scholar], [Publisher]
- Choudhury N R, Malik P S, Singh D K, Islam M N, Kaliappan K, Mukherjee S K. (2006). The oligomeric Rep protein of Mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic acids research, 34(21): 6362-6377. Crossref], [Google Scholar], [Publisher]
- Wang D, Zhang X, Yao X, Zhang P, Fang R, Ye J. (2020). A 7-amino-acid motif of Rep protein essential for virulence is critical for triggering host defense against Sri Lankan cassava mosaic virus. Molecular Plant-Microbe Interactions, 33(1): 78-86. [Crossref], [Google Scholar], [Publisher]
- George B, Ruhel R, Mazumder M, Sharma V K, Jain S K, Gourinath S, Chakraborty S. (2014). Mutational analysis of the helicase domain of a replication initiator protein reveals critical roles of Lys 272 of the B′ motif and Lys 289 of the β-hairpin loop in geminivirus replication. Journal of General Virology, 95(7): 1591-1602. [Crossref], [Google Scholar], [Publisher]
- Settlage S B, See R G, Hanley-Bowdoin L. (2005). Geminivirus C3 protein: replication enhancement and protein interactions. Journal of virology, 79(15): 9885. [Crossref], [Google Scholar], [Publisher]
- Selth L A, Dogra S C, Rasheed M S, Healy H, Randles J W, Rezaian M A. (2005). A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. The Plant Cell, 17(1): 311-325. [Crossref], [Google Scholar], [Publisher]
- Kass G, Arad G, Rosenbluh J, Gafni Y, Graessmann A, Rojas M R, Gilbertson R L, Loyter A. (2006). Permeabilized mammalian cells as an experimental system for nuclear import of geminiviral karyophilic proteins and of synthetic peptides derived from their nuclear localization signal regions. Journal of general virology, 87(9): 2709-2720. [Crossref], [Google Scholar], [Publisher]
- Sanderfoot A A, Ingham D J, Lazarowitz S G. (1996). A Viral Movement Protein as a Nuclear Shuttle (The Geminivirus BR1 Movement Protein Contains Domains Essential for Interaction with BL1 and Nuclear Localization). Plant physiology, 110(1): 23-33. [Crossref], [Google Scholar], [Publisher]
- Zhang S, Ghosh R, Jeske H. (2002). Subcellular targeting domains of Abutilon mosaic geminivirus movement protein BC1. Archives of virology, 147(12): 2349-2363. [Crossref], [Google Scholar], [Publisher]
- Betts M J, Russell R B. (2003). Amino acid properties and consequences of substitutions. Bioinformatics for geneticists, 317(289): 10.1002. [Crossref], [Google Scholar], [Publisher]
- Sanz A I, Fraile A, García-Arenal F, Zhou X, Robinson D J, Khalid S, Butt T, Harrison B D. (2000). Multiple infection, recombination and genome relationships among begomovirus isolates found in cotton and other plants in Pakistan. Microbiology, 81(7): 1839-1849. [Crossref], [Google Scholar], [Publisher]
- Sánchez-Campos S, Domínguez-Huerta G, Díaz-Martínez L, Tomás D M, Navas-Castillo J, Moriones E, Grande-Pérez A. (2018). Differential shape of geminivirus mutant spectra across cultivated and wild hosts with invariant viral consensus sequences. Frontiers in plant science, 9: 932. [Crossref], [Google Scholar], [Publisher]
- Aimone C D, Lavington E, Steen Hoyer J, Deppong D O, Mickelson-Young L, Jacobson A, Kennedy G G, Carbone I, Hanley-Bowdoin L, Duffy S. (2021). Population diversity of cassava mosaic begomoviruses increases over the course of serial vegetative propagation. J Gen Virol., 102(7): 001622. [Crossref], [Google Scholar], [Publisher]
- Domingo E, García-Crespo C, Perales C. (2021). Historical Perspective on the Discovery of the Quasispecies Concept. Annual Review of Virology, 8: 51–72. [Crossref], [Google Scholar], [Publisher]
- Pooggin M M. (2017). RNAi-mediated resistance to viruses: a critical assessment of methodologies. Current opinion in virology, 26: 28-35. [Crossref], [Google Scholar], [Publisher]